ASAP’s mission is to accelerate the pace of discovery and inform the path to a cure for Parkinson’s disease (PD) through collaboration, research-enabling resources, and data sharing. The year 2021 proved to be one of significant momentum and impact for our initiative, our network, and the ASAP-supported resource projects that are fueling research, enabling studies, and nurturing the entire neuroscience community.
Research Outputs Across ASAP Programs
Collaborative Research Network
Added 14 new teams to the network.
Published 24 original research, reviews, and op-eds.
Global Parkinson’s Genetics Program
- Established the first biobank in Peru.
- Expanded global reach by launching GP2 website in 6 additional languages.
Parkinson’s Progression Markers Initiative
- Publicly relaunched the study with in-clinic recruitment and online platform.
- Continued to grow a robust, open-access database with the addition of functional omics data generated from 95 iPSC lines.
Accelerating Medicines Partnership® in Parkinson’s Disease
- Shared 3 GP2 datasets within AMP® PD comprising over 4800 individual PD and matching control cases.
iPSC Neurodegenerative Disease Initiative for PD
- Launched iNDI-PD to develop a PD shared resource for iPSC cell lines.
Open Science
- Remained active member of open science community, joined the ORFG, and spoke at different events.
- Partnered with DataSeer.Ai to develop a tool to promote best practices in open science.

The ASAP Collaborative Research Network is an international, multidisciplinary, and multi-institutional network of 35 teams working to address research and knowledge gaps in the development and progression of PD.
Expanded the CRN with New Teams of Grantees
This year, the CRN welcomed 14 new research teams under a circuitry and brain-body interactions theme. These teams will focus their research on understanding how the circuits that underlie key brain regions are affected in PD and how they may contribute to disease initiation and progression.

Published Novel Discoveries
In our commitment to open science, our teams share their discoveries early, posting their findings to preprint servers. Check out our bioRxiv/medRxiv preprint channel for more details.
This year, our two funded themes – neuro-immune and functional genomics – published 9 reviews and 15 original research open-access papers in various journals.
The research discoveries largely centered around 4 categories:
1. Selective Autophagy
Dysfunction of selective autophagic mechanisms have been directly linked to PD. ASAP teams published articles that centered on understanding autophagosome biogenesis, elongation, and downstream mechanisms, with specific insights on mitophagy and lysophagy.
- Global ubiquitylation analysis of mitochondria in primary neurons identifies endogenous Parkin targets following activation of PINK1 (Teams Alessi & Harper Collaboration)
- VPS13D bridges the ER to mitochondria and peroxisomes via Miro (Team De Camilli)
- Quantitative proteomics reveals the selectivity of ubiquitin-binding autophagy receptors in the turnover of damaged lysosomes by lysophagy (Team Harper)
- Structural basis for membrane recruitment of ATG16L1 by WIPI2 in autophagy (Team Hurley)
- Reconstitution of cargo-induced LC3 lipidation in mammalian selective autophagy (Team Hurley)
- ALS- and FTD-associated missense mutations in TBK1 differentially disrupt mitophagy (Team Hurley)
2. Alpha-Synuclein and Tau Pathology
Protein aggregation has been implicated in the progression of neurodegenerative disorders, such as PD. Focusing specifically on the proteins alpha-synuclein and tau, ASAP teams explored how protein aggregation begins and how it can be hindered or enhanced.
3. LRRK2 Biology
LRRK2 mutations are the most common genetic causes for PD. ASAP teams focused on understanding the LRRK2 interactome and how LRRK2 mutations impact downstream signaling pathways.
4. Comparative Analysis Between Parkinson’s Disease and Other Diseases
Diseases are often complex and multi-systemic, and as such can have shared pathophysiology. ASAP teams explored PD in the context of other diseases and created tools to better evaluate risk loci.
- Parkinson’s disease and cancer: a systematic review and meta-analysis of over 17 million participants (Team Chen)
- Integrating protein networks and machine learning for disease stratification in the Hereditary Spastic Paraplegias (Team Hardy)
- powerEQTL: an R package and shiny application for sample size and power calculation of bulk tissue and single-cell eQTL analysis (Team Scherzer)
Generated Resources
ASAP-funded resources generated by the CRN.
50
Plasmids
15
Protocols
12
Datasets
5
Code/Software Tools
Hosted Our First Annual Celebration of Scientific Achievement (COSA) Virtual Event
This event was designed to showcase the unique contributions of young investigators across the CRN. Over 75 abstracts were submitted, and 267 individuals registered from across our network. Three Community Prizes (voted by all attendees) and three Reviewer Prizes (voted by reviewers assessing the presentations) were awarded.

COSA community prize winners:

Daniel Saarela (Team Alessi)
Rapid Isolation of Organelles to Study LRRK2 Signaling

Rotimi Fasimoye (Team Alessi)
GOLGI-IP: A Novel Tool for Multimodal Analysis of Golgi Molecular Content

Stephanie Vrijsen (Team Vangheluwe)
Sequential Lysosomal and Mitochondrial Immunoprecipitation for Metabolomics Analysis
COSA reviewer prize winners:

Kerryn Berndsen (Team Alessi)
For her presentation on Structural Basis for the Specificity of RAB Recognition by PPM1H Phosphatase

Edmundo Vides (Team Alessi)
For his presentation on Molecular Analysis of LRRK2 Membrane Recruitment

Angelique di Domenico (Team Studer)
For her presentation on Intrinsic LRRK2 Parkinson’s Disease Phenotypes using Patient Specific iPSC-derived Models
This year is only the beginning! We look forward to seeing how the network grows over the next year, in particular with our newly funded Circuitry theme that will focus on understanding the neuronal circuitry of Parkinson’s disease.

GP2 aims to genotype the most ancestrally diverse cohort of PD participants in the world. In its second year, GP2 is making strides in PD research and knowledge sharing.
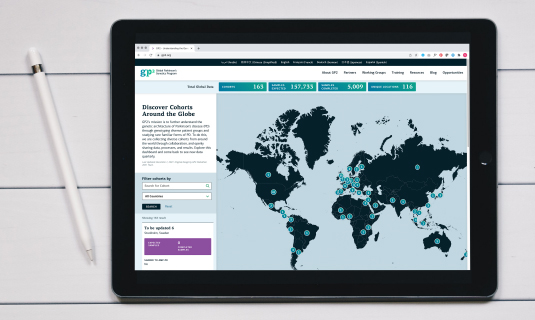
Developed Cohort Dashboard to Illustrate Genotyping Progress
GP2 continues to collect diverse cohorts from around the world through collaboration and openly sharing data, processes, and results. Explore the dashboard and learn more about our cohort collaborators.
Expanded Global Reach with Language Accessibility
GP2 launched its site in six new languages this year, as part of its commitment to broaden the reach of resources, training, and opportunities and to expand global collaboration and open data around the world. GP2.org is now available in Arabic, Chinese, French, German, Japanese, and Spanish.

Expanded Opportunities for Researchers
Creating a global generation of scientists and providing opportunities for groups that are traditionally underrepresented in research are key outcomes for GP2. This year, GP2 supported four new Ph.D. trainees whose research will help to increase Asian and Latin American representation in the PD field. Learn more about them here.
Launched Online Training Platform
Training is a foundational element of GP2. This year, GP2 launched its online Rise training platform, making core coursework accessible to the neurodegenerative research community. Developed by GP2’s Training, Networking, and Communication Working Group, Rise features course offerings focused on data analysis, bioinformatics, and genetics, and a curated video series. Explore and register for the Rise platform.
Launched Monogenic Hub Website and Portal
GP2’s new Monogenic Hub website offers access to insights across working groups and explores the goals of each team within GP2. The Monogenic Portal provides a platform through which researchers can submit patient families and cases. The Portal is also a place for researchers to share existing clinical and genetic data within the Monogenic Hub. Cases submitted through the Portal are evaluated and potentially prioritized for whole-genome sequencing. GP2 continues to make significant strides towards its goal of sequencing >10,000 individuals.


Introduced BLAAC PD Initiative
Around 90% of genetic studies in the PD field have been performed on European ancestry populations. In our commitment to close the gap in this disparity, GP2 launched the Black and African American Connections to Parkinson’s Disease (BLAAC PD) study which will investigate the genetic impact on PD susceptibility specific to these populations. Learn more.
Opened First-of-its-kind Biobank in Peru
Peru’s Instituto Nacional de Ciencias Neurológicas celebrated the opening of its DNA biobank, the first of its kind in Peru. The biobank will serve as a repository for genetic samples, implementing a system and standards for the use of genetic information for research at the institute and other national and international collaborators.

Showcased Thought Leadership
Connecting with others in the community: In June, GP2 held its bi-annual meeting to connect the GP2 network with interested researchers, provide updates on the program’s progress to date, and discuss opportunities to address research barriers identified in the community. The two-day virtual meeting hosted over 200 participants from around the world.
Promoting ancestral diversity in genetic studies more broadly: In October, our Managing Director, Ekemini Riley, presented at the National Academy of Science, Engineering and Medicine (NASEM) on “Molecular insights to patient stratification for neurological and psychiatric disorders,” showcasing how others could leverage the process laid out by GP2 to ensure that underrepresented populations are represented in genetic research studies. In November, Ekemini co-authored an op-ed in Nature Cell Biology discussing the need for incorporating diversity into basic science models as well.
Shared Tools
In addition to its commitment to sharing data sets generated from this program, GP2 is also committed to open science best practices, sharing a suite of tools for processing genetic data that are accessible and free to all researchers.

PPMI is a groundbreaking study sponsored by The Michael J. Fox Foundation, to provide deeper understanding of the biological and clinical underpinnings of PD, working with partners worldwide, to build a solid open-access data collection and biosample library to accelerate scientific breakthroughs and innovative therapies.
Demonstrated Reproducible High Diagnostic Performance of Protein Seeding Assays
PPMI scientists — supported by MJFF and PPMI study partners including ASAP — compared alpha-synuclein seed amplification assay results obtained from three independent laboratories to evaluate reproducibility across methodological variations. Results were remarkably similar across laboratories, displaying high diagnostic performance (sensitivity ranging from 86 to 96% and specificity from 93 to 100%) and further validating the diagnostic potential of seed amplification assays for well-characterized early de novo PD. Learn more.
Shared Functional Omics Data
The multi-institutional FOUNdational Data INitiative for Parkinson’s Disease (FOUNDIN-PD) project led by GP2 principal investigator Andrew Singleton, PhD, has released functional omics data from dopamine neurons derived from 95 PPMI iPSC lines and generated cellular imaging data from iPSC-derived dopamine neurons to understand key molecular relationships in a PD-relevant cell-type. PPMI generated ‘omics data on biosamples to complement other biological, clinical, and imaging data to enable unprecedented comparison studies. View tables and data from FOUNDIN-PD analysis.

Launched Ambitious In-Clinic Expansion and Online Platform
With ASAP support, PPMI has launched a seismic expansion to more than double in-clinic recruitment (including 2,000 prodromal volunteers). To aid recruitment, an online study platform opened in July 2021 to anyone over 18 in the U.S. This tool gathers data over time on health and wellness and identifies individuals with certain risk factors who may be eligible for in-clinic participation. Thus far, more than 75,000 people have taken an online action to join PPMI.
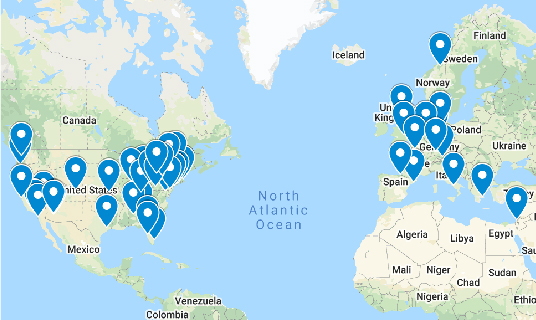
Expanded into More Countries with New Sites
The PPMI expansion added 17 new clinical sites and two new countries (Canada and the Netherlands) to its roster of medical centers enrolling and following participants. Now at 49 sites in 12 countries — with plans to announce another new site in early 2022 — the study is broadening its global reach and, with it, the picture of Parkinson’s disease.

Shared Data and Resources Streamlined with New Website and Partnerships
The PPMI open-access data set has been downloaded over 8.5 million times. These data and available biosamples are valued resources for the community. In 2021, PPMI added raw and “derived” data from the Verily Study Watch, and launched a new researcher-facing website to more clearly display data dashboards, dictionaries, publications, and presentations, to encourage use of resources and impact of findings on future projects.
PPMI data also flows to GP2 and is harmonized and shared through the AMP PD program.

The iPSC Neurodegenerative Disease Initiative (iNDI) aims to understand better how genetic abnormalities cause the brain cell damage that causes Alzheimer's disease and other dementias. ASAP is supporting iNDI-PD, an expansion to ensure that additional mutations relevant to PD research are also included in this effort.

Program Launch
iNDI-PD has already made great strides in providing materials for the PD scientific community. To date, 19 human isogenic iPSC lines expressing PD-associated mutations (concentrated on mutations in 6 different genes) have already been developed. These iPSC lines will begin to be available from JAX labs as early as the first quarter of 2022. Learn more about the initiative.
The Accelerating Medicines Partnership (AMP) is a public-private partnership between the National Institutes of Health (NIH), multiple biopharmaceutical and life sciences companies, and non-profit organizations.

GP2 and AMP PD Partner to Become a One-Stop Shop for PD Data
GP2 and AMP PD have partnered to be a one-stop shop for Parkinson’s disease genetic (GP2) and genomic (AMP PD) data, allowing researchers to find new drug targets and biomarkers faster and develop new treatments faster. Additional highlights include:
- The first GP2 data-release within AMP PD (established the partnership between GP2 and AMP PD).
- The integration of the first batch of GP2 data, which allows users to analyze 4874 samples across 3 cohorts, thus increasing the power of follow-on analysis.

By supporting open science, we facilitate the rapid and free exchange of scientific ideas, ensuring that the research we fund can be leveraged for future discoveries. Open science is at the heart of ASAP’s mission and work.
Joined the Open Research Funders Group (ORFG)
The ORFG is a coalition of leading philanthropic organizations committed to supporting the open sharing of research to benefit scientific progress. Membership will continue to optimize our efforts to foster collaboration within its network, tackle field-wide challenges, and accelerate the discovery of a PD cure.
Partnered with DataSeer
In partnering with AI-powered service provider DataSeer, we co-created a tool to help guide our network in following best practices in open science. As an AI-powered service provider, DataSeer leverages artificial intelligence to ensure all manuscripts submitted for journal peer review by ASAP grantees include associated datasets, code, protocols, and lab materials.

Actively Engaged with the Broader Open Science Community
ASAP is an active member in the open science community. This year, our very own Deputy Director, Sonya Dumanis, spoke at three events:
1) the International Network for Government Science Advice (INGSA) Satellite event dedicated to science in the 21st century.
2) a panel at the Research Data Alliance’s 17th Plenary Meeting on “Delimiting the units of open data.”
3) Openscapes Champions Program; Symbiosis in Aquatic Systems Initiative (SASI) Cohort, supported by the Moore Foundation and co-led with Protocols.io.
We also have representatives sitting on cOAlitionS subcommittees and the ORFG.
Onward!
As we reflect on our outcomes this year, Aligning Science Across Parkinson's (ASAP) thanks our funders, partners, research community, and stakeholders for every contribution they have made to advance collaboration, creativity, flexibility, and transparency in accelerating and informing the path to a cure for Parkinson’s disease. We know that 2022 will be full of more discoveries and accomplishments. We look forward to our continued partnership and support to strengthen our collaborative research community efforts.

