As we reflect on the second full year of Aligning Science Across Parkinson’s (ASAP), our initiative has accomplished a tremendous amount. We are proud of the impact our initiative, our network, and ASAP-supported programs are having on the Parkinson’s disease community.
OUR VISION:
Collaborative and transparent research processes and environments that deliver faster and better outcomes for Parkinson’s disease.
OUR MISSION:
Accelerating the pace of discovery and informing the path to a cure for Parkinson’s disease through collaboration, research-enabling resources, and data sharing.
ASAP’s Theory of Change for Open and Collaborative Science
ASAP applies a Theory of Change framework, which allows us to cohesively and comprehensively measure progress – from idea to result – in the context of our overarching vision. Further, it enables us to reflect upon and iterate processes as we measure our progress to date. Our work is broken down into three main components, or goals, that drive our mission forward:
SUPPORT COLLABORATION
Scientific progress can be accelerated when researchers exchange ideas early and often, in a collaborative rather than competitive manner.
GENERATE RESOURCES
By supporting resource development, we are building an infrastructure, available to the larger scientific community, that improves access to research tools, reproducibility of studies, and process efficiency to accelerate discoveries.
SHARE DATA
By sharing research outputs like data, code, and protocols, we are allowing researchers to build upon the work of others. This facilitates collaboration among investigators, attracts new talent and expertise to the field, and allows us to increase the power of our studies through meta-analysis.
ACCELERATE DISCOVERIES
We believe that by supporting collaboration, facilitating research, enabling resource generation, and creating a culture of data sharing, we can deliver faster and better outcomes for Parkinson’s disease research.
Program Specific Overview & Milestones
In the following sections, we highlight some of the 2022 outputs from ASAP’s supported programs and show how these outputs inform our goals within the Theory of Change framework.
Collaborative Research Network (CRN)
Global Parkinson’s Genetics Program (GP2)
Parkinson’s Progression Markers Initiative (PPMI)
IPSC Neurodegenerative Diseases Initiative (iNDI-PD)
Accelerating Medicine Partnership in Parkinsons Disease (AMP® PD)

The ASAP Collaborative Research Network (CRN) is an international, multidisciplinary, and multi-institutional network of 35 teams working to address research and knowledge gaps in the development and progression of Parkinson's disease (PD).
The CRN was designed to create an environment that facilitates the rapid and free exchange of scientific ideas that would spark new discoveries for PD.
SUPPORT COLLABORATION
Held First Annual In-Person Meeting
More than 200 members of the CRN gathered in April for the inaugural annual in-person meeting that was designed to echo ASAP’s mission to foster collaboration, encourage data sharing, and support resource generation. In a post-event survey, 100% of respondents reported meeting someone they had never met, and 78% mentioned that their own research would be reshaped based on connections they made during the meeting. Read more about the event.

“I feel like I have much more breadth and depth of understanding of different aspects of current research, and it was really great to be able to network and meet people on other teams, to talk synergy, and to learn from each other.”
–Samantha Gruenheld, PhD, Team Desjardins, McGill University
SUPPORT COLLABORATION
Hosted the Celebration of Scientific Achievement (COSA) Event
COSA 2022 had over 440 individuals registered to attend from across the ASAP network, with over 140 abstracts showcased. From our poster presentations, five presenters received awards for their outstanding contributions to the PD field.

COSA Prize Winners

FIRST PLACE
Miguel Chuapoco (Team Gradinaru)
Intravenous Gene Transfer Throughout the Brain of Infant Old World Primates Using Adeno-Associated Virus (AAV)

Eduard Bentea (Team Vangheluwe)
Exploring the Role of the Lysosomal Protein ATP10B in the Nigrostriatal Dopaminergic Pathway of Rats

SECOND PLACE
Natalie Kaempf (Team Voet)
Sleep Defects Delineate Parkinson’s Disease Subgroups in Drosophila

Hariam Raji (Team Schapira)
Long-Term Culture and Characterization of GBA1 Midbrain Organoids

Sherilyn Junelle Recinto (Team Desjardins)
Dysregulated Immune Response in the Gut Underlying Early Parkinson’s Disease Symptoms
SUPPORT COLLABORATION/SHARE DATA
Hosted Virtual Events
Throughout the year, members of the CRN have met virtually to discuss findings around a variety of topics. Of the 30+ virtual special interest group events in 2022, over 94% of them shared preliminary data during the meeting.
GENERATE RESOURCES/SHARE DATA
Shared Resources
We shared over 1,500 resources within our network, showcasing the research outputs and tools used by all ASAP-funded programs regardless of funding source. All the ASAP-funded work is also cataloged online. We have over 400 items, from protocols to code to datasets, and are adding more each month.

GENERATE RESOURCES
Launched Additional Resource Support Programs
We launched two additional resource support programs.
- Alpha-Synuclein Preformed Fibril Core
As many in our network are using alpha-synuclein preformed fibrils (PFF) to generate their PD-relevant models, we launched a preformed fibrils resource core to ensure standardization and reproducibility of results across teams. The purpose of this resource team is to provide the ASAP network with consultation and access to standardized PFFs as needed, to lead workshops on PFF-related techniques, and to support the development of a PFF model at a contract research organization for future studies that could benefit those both within and outside the CRN.
- Interrogating the Gut to Brain Theory of PD
There is a debate in the PD field about whether alpha-synuclein aggregation starts in the gut or in the brain. Last year, ASAP members Thomas Beach and Geidy Serrano published the results of an extensive autopsy investigation looking at the presence of abnormal synuclein in the stomach and vagus nerve and only observed pathology there when brain pathology was also present. This finding suggests that the body-first theory may be incorrect. To settle this debate and aid ASAP teams exploring this theory, ASAP is supporting an extension of their work to explore if other conformers of alpha-synuclein are present in 200 autopsied individuals by using a more sensitive and specific method to detect abnormal alpha-synuclein aggregation. The data from this study will be available online and the tissues examined will be available to any researchers upon request once the study is complete.
ACCELERATE DISCOVERIES
Sharing Findings Earlier
In accordance with our commitment to open science, CRN grantees post preprints an average of six months before they are released for final publication. As of December 6, 2022, we supported 75 research articles, of which 36 have completed peer review and been released for final publication. Research articles shared findings from our network across disciplines, ranging from Neuroscience to Cell Biology to Immunology. Our network also posted 34 reviews showcasing thought leadership around emerging topics from Neuroscience to Microbiology to Genomics.
Learn more about the articles – including preprints, publications, and reviews – by looking at our ASAP Catalog. You can also stay up to date on the latest ASAP-funded preprints and the latest published articles.
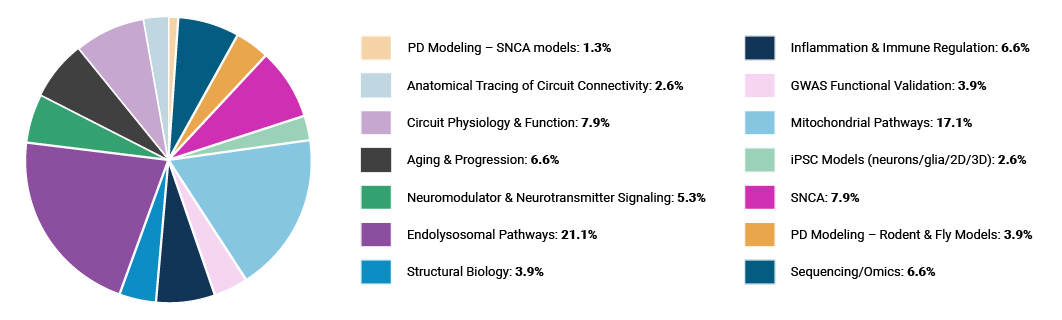
As 2022 comes to a close, the CRN community – of over 1,000 researchers across the globe at various stages of their professional careers – is committed to facilitating the rapid and free exchange of scientific discoveries to spark new discoveries for PD. We developed a directory to highlight the over 200 CRN Investigators leading ASAP-funded research projects across our 35 teams, as well as the work they are doing and their scientific contributions to the PD field.

GP2 aims to understand the genetic architecture of PD and make that knowledge globally relevant. To this end, GP2 is working toward three main goals: harmonize existing data collections and support ongoing initiatives around the world; analyze and understand ancestral significance; and build local capacity within underrepresented regions.
To ensure that ancestral diversity information was being included in PD-related genetic datasets around the globe, GP2 set up infrastructure to centralize this information and allow these datasets to be continually analyzed, thereby increasing our understanding of PD across populations.
SUPPORT COLLABORATION
Harmonizing Existing Efforts and Supporting Ongoing Initiatives
64 organizations from 57 regions around the world have raised their hands to be part of the GP2 effort. As of December 6, 2022, there are a total of 136 cohorts involved in the program across 199 unique sites. Submit a cohort by emailing [email protected].

SUPPORT COLLABORATION
Hosted First Annual In-Person GP2 Meeting
More than 170 participants from 49 countries met in September for the first in-person GP2 meeting. At the gathering, GP2’s core tenets of collaboration and openness of data, methods, and results were integral as attendees discussed progress across the program’s areas of focus and mapped the future. A highlight of the meeting was hearing from the next generation of PD genetic researchers serving underrepresented populations. Check out our recap and watch our video to meet a few of the attendees.

GENERATE RESOURCES
Building Local Capacity
We are expanding PD research within underrepresented regions and establishing a strong and diverse global pipeline of PD researchers. In 2022, we:
- Launched a virtual meeting series of 100+ trainees within the GP2 network who meet on a monthly basis.
- Awarded trainee grants to two new Masters’s students from Djibouti and Ghana.
- Developed over 70 virtual courses on the Rise platform, with subtitles available in over 100 languages that 700+ individuals are currently utilizing.
GENERATE RESOURCES
Developing the Infrastructure to Promote PD Genetics Research
In 2022, we:
- Launched the Monogenic Resource Map to facilitate research exchange and share clinical trial information around monogenic forms of PD.
- Leveraged GP2 infrastructure to support new PD genetics initiatives for Black and African Americans, U.S. veterans, individuals from Central Asia, and diverse populations from Australia.
- Hosted an IPDGC/GP2 virtual hackathon with researchers across 12 countries to develop novel tools for analyzing PD genetic datasets. Learn more about the event by reading our preprint.
SHARE DATA
Continuing Our Commitment to Transparency
Underlying raw data and summary statistics from our global cohorts are now available for over 14,900 samples around the world through the AMP® PD portal.
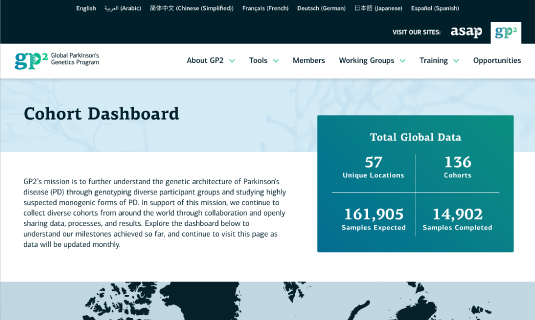
We revamped the cohort dashboard to better showcase our progress and completed three data releases in 2022. Explore our updated cohort dashboard.
ACCELERATE DISCOVERIES
Analyzing and Understanding Ancestral Significance in PD Genetics Research
GP2 conducted the first large-scale, multi-ancestry meta-analysis of PD with 49k+ cases, 18k+ proxy cases, and 2 million+ controls among those with European, East Asian, Latin American, and African ancestry. Twelve (12) potentially novel loci were identified in non-European populations.
As we wrap up our efforts in 2022, GP2 maintains its commitment to global collaboration in making progress toward its three goals. To this end, we have extended our support for GP2 from a five-year to a ten-year-long initiative. Visit our working group directory to engage with our team.

Parkinson’s Progression Markers Initiative (PPMI) is a groundbreaking study sponsored by The Michael J. Fox Foundation to provide a deeper understanding of the biological and clinical underpinnings of PD. We work with partners worldwide to build a solid open-access data collection and biosample library to accelerate scientific breakthroughs and innovative therapies.
Through PPMI, ASAP is proud to participate alongside other PPMI study partners in efforts to develop worldwide comprehensive strategies for investigating and defining biomarker signatures for those in the very early stages of PD.
SUPPORT COLLABORATION
Expanding the Global Network
In 2022, PPMI has achieved the following:
- Four (4) additional clinical sites joined the existing 49 activated international sites, with the first African PPMI site activated in Lagos, Nigeria.
- PPMI online has begun recruiting outside of the United States, collaborating with the following international online outreach programs:
- Healthy Brain Initiative in Germany, Spain, Austria, and Luxembourg
- Israel Prodromal Parkinson Initiative (IPPI) in Israel
- EPIPARK in Germany
- PPMI expanded enrollment of participants with REM sleep behavioral disorder (RBD) into PPMI at 19 clinical sites to help identify individuals at higher risk of developing PD.
GENERATE RESOURCES
Increased Participation in Studies
PPMI’s 2022 efforts have led to increased participation in PD studies:
- Enrolled over 30,000 individuals in PPMI online this year.
- Developed a novel smell test initiative to expand prodromal enrollment worldwide with over 10,000 participants participating. Learn more at Smell Test Direct.
- Fielded over 50 biosample collection share requests in 2022.
- Expanded the database by including neuropathy autopsy results from PPMI participants.
SHARE DATA
Increasing Data Usage
PPMI has continued to prioritize data sharing this year.
- PPMI data has been downloaded over 10 million times. In the past year, there were more than 1 million downloads.
- To share discoveries earlier from PPMI cohorts, the PPMI Zenodo community was established to share early findings presented at conferences.

ACCELERATE DISCOVERIES
Made New Findings
PPMI has made the following discoveries in 2022:
- Tested novel molecular imaging tracers in PPMI participants to identify better PET biomarkers for PD onset and progression.
- Supported algorithm development incorporating different biomarkers (such as clinical olfactory function, DAT imaging, and the synuclein seeding assay) to identify subsets of Parkinson’s phenotypes.
- Developed a new biological PD staging hypothesis based on the results of the synuclein seeding assay in CSF samples collected from over 1,000 individuals in PPMI, which has implications for future clinical trial enrollment planning.
- Discussed with regulators how to establish a therapeutic prevention platform master protocol for conducting therapeutic trials in at-risk individuals.
- Conducted genetic analysis in individuals enrolled in PPMI who have been diagnosed with PD by a clinician but seem to have normal dopaminergic functional imaging scans suggesting this was a misdiagnosis. Findings suggest that hereditary dystonia may be a significant contributing factor to the misdiagnosis of PD. See the preprint.

We are grateful for our collaboration with The Michael J. Fox Foundation and PPMI to better define and measure PD to speed therapeutic development. By supporting the seismic expansion of PPMI, the initiative is now able to better understand the early stages of PD and the resulting progression across the population.

The iPSC Neurodegenerative Disease Initiative (iNDI) is the largest genome engineering initiative in research to date anchored within the NIH Center for Alzheimer’s and Related Dementias (CARD). ASAP supports iNDI-PD, an extension of the iNDI research initiative that draws from induced pluripotent stem cells (iPSCs), to include PD in its research to discover novel insights into the molecular biology of disease and to ensure that additional mutations relevant to Parkinson’s disease research are also included in their efforts.
Our support of iNDI-PD is designed to ensure that there are accessible iPSC resources for all researchers interested in understanding PD. By incorporating PD data in the neurodegenerative resource bank, we are also enabling researchers to draw connections across diagnostic lines.
SUPPORT COLLABORATION
Piloting the Use of Early-Release Cell Lines
As cell lines get developed, iNDI is providing members of our Collaborative Research Network the opportunity to beta-test cell lines while they undergo extensive quality control. All cell lines will be made available to all researchers on the distribution website listed above once quality control has been completed.
Image from Agnete Kirkeby, Kirkeby Lab, University of Copenhagen
GENERATE RESOURCES
Established KOLF2.1J as a Cell Reference Line and Made Data on the Line Widely Available
To select a reference cell line, the iNDI team tested a variety of iPSC-derived lines, deeply characterized their genetic properties, assessed the genomic stability of the subclones, and tested their performance with labs around the world. The findings and associated data, such as sequencing and code, have been freely shared for other researchers to leverage in their iPSC studies.
GENERATE RESOURCES
Launched the Distribution Website
This website allows researchers to order the specifically engineered lines designed by the iNDI-PD team to study genetic variants behind neurodegenerative disease with a click of a button.
We are proud to support iNDI-PD and ensure that additional genetic mutations relevant to PD research are included in this effort. As the iNDI-PD resources become available, we want researchers to be able to use them as needed. If you are interested in receiving alerts as new lines become available:

The Accelerating Medicine Partnership® in Parkinson’s Disease (AMP® PD) is a public-private partnership between the National Institutes of Health (NIH), multiple biopharmaceutical and life science companies, and nonprofit organizations with the goal of creating a large harmonized dataset to advance the development of Parkinson’s disease therapies.
ASAP is proud to be a partner in the AMP® PD effort to broaden data sharing in the biomedical community to advance PD research. Both GP2 and PPMI cohorts share their data on this platform.
GENERATE RESOURCES
Committing to the User Experience
- AMP® PD has launched its first Community Workspace on the AMP® PD Knowledge Platform, the AMP-PD RNA-Seq Explorer, created by David Craig’s laboratory at USC.
- Three new Terra curated Jupyter notebooks and feature workspaces were created and provided for AMP® PD users: Getting Started Proteomics, AMP® PD – Proteomics QC and Analysis, and AMP® PD – Polygenic Risk Scores.
- New R Markdown analyses were provided for users within Terra facilitating the use of R Studio for the analysis of AMP® PD data.
SHARE DATA
Expanding Access to GP2 Data on the AMP® PD Platform
Throughout 2022, GP2 collaborated with AMP® PD to continue releasing PD genetic data for research of novel drug targets and biomarkers through collaborative cloud-based computing. The AMP® PD cloud platform provides a single secure access point for data collated and generated by GP2 to prioritize the safety and security of data through verified researcher authentication. Future data releases will continue to grow the diversity of participants available. Learn how to apply for data access.
- AMP® PD’s updated version 2.5 public dataset includes adjusted genetic mutation and variant (i.e., APOE status) information, as well as additional targeted proteomics metadata and QC information.
- RNA sequencing data from extracellular vesicles were added to the platform.
- Additional Olink targeted proteomics were sponsored by Abbvie for addition to the platform.
- There are over 725 users registered with AMP® PD to access the data.
ACCELERATE DISCOVERIES
Citing AMP® PD Data in Publications
Eleven (11) publications cited AMP® PD data over the last year. Read more below:
Eric B. Dammer, Lingyan Ping, Duc M. Duong, Erica S. Modeste, Nicholas T. Seyfried, James J. Lah, Allan I. Levey, Erik C.B. Johnson.
Junfeng Luo, MS, Hao Wu, BS, Jinxia Li, BS, Wenbiao Xian, MD, Weimin Li, MS, Joseph J. Locascio, PhD, Zhong Pei, MD, Ganqiang Liu, PhD*.
William Zhu, Xiaoping Huang, Esther Yoon, Sara Bandres-Ciga, Cornelis Blauwendraat, Kimberly J Billingsley, Joshua H Cade, Beverly P Wu, Victoria H Williams, Alice B Schindler, Janet Brooks, J Raphael Gibbs, Dena G Hernandez, Debra Ehrlich, Andrew B Singleton, and Derek P Narendra.
Erinc Hallacli, Can Kayatekin, Sumaiya Nazeen, Xiou H. Wang, Zoe Sheinkopf, Shubhangi Sathyakumar, Souvarish Sarkar, Xin Jiang, Xianjun Dong, Roberto DiMaio, Wen Wang, Matthew T. Keeney, Daniel Felsky, Jackson Sandoe, Aazam Vahdatshoar, Namrata D. Udeshi, DR Mani, Steven A. Carr, Susan Lindquist, Philip L. de Jager, David P. Bartel, Chad L. Myers, Timothy J. Greenamyre, Mel B. Feany, Shamil Sunyaev, Chee Yeun Chung, and Vikram Khurana.
Cell 2022, 185(12): 2035-2056.e33.
KR Bowles, DA Pugh, Y Liu, T Patel, AE Renton, S Bandres-Ciga, Z Gan-Or, Heutink, A Siitonen, S Bertelsen, JD Cherry, CM Karch, SJ Frucht, BH Kopell, I Peter, YJ Park, International Parkinson’s Disease Genomics Consortium (IPDGC), A Charney, T Raj, JF Crary, AM Goate.
Raquel Real, Alejandro Martinez-Carrasco, Regina H. Reynolds, Michael A. Lawton, Manuela M. X. Tan, Maryam Shoai, Jean-Christophe Corvol, Mina Ryten, Michele T. M. Hu, Yoav Ben-Shlomo, Donald G. Grosset, John Hardy, Huw R. Morris*.
Bernabe I Bustos, Sara Bandres-Ciga, J Raphael Gibbs, Dimitri Krainc, Niccolo E Mencacci, Ziv Gan-Or, Steven J Lubbe, International Parkinson’s Disease Genomics Consortium (IPDGC).
Zhenhua Liu*, Nannan Yang*, Jie Dong*, Wotu Tian, Lisa Chang, Jinghong Ma, Jifeng Guo, Jieqiong Tan, Ao Dong, Kaikai He, Jingheng Zhou, Resat Cinar, Junbing Wu, Armando Salinas, Lixin Sun, Mantosh Kumar, Breanna Sullivan, Braden Oldham, Vanessa Pitz, Mary Makarious, Jinhui Ding, Justin Kung, Chengsong Xie, Sarah Hawes, Lupeng Wang, Tao Wang, Piu Chan, Zhuohua Zhang, Weidong Le, Shengdi Chen, David M. Lovinger, Cornelis Blauwendraat, Andrew Singleton, Guohong Cui, Yulong Li, Huaibin Cai, and Beisha Tang.
Francis P. Grenn, Mary B Makarious, Sara Bandres-Ciga, Hirotaka Iwaki, Andrew Singleton, Mike Nalls, Cornelis Blauwendraat, The International Parkinson Disease Genomics Consortium.
Steven J Lubbe, Bernabe I Bustos, Jing Hu, Dimitri Krainc, Theresita Joseph, Jason Hehir, Manuela Tan, Weijia Zhang, Valentina Escott-Price, Nigel M Williams, Cornelis Blauwendraat, Andrew B Singleton, Huw R Morris, and for International Parkinson’s Disease Genomics Consortium (IPDGC).
Paolo Reho, Shunsuke Koga, Zalak Shah, Ruth Chia, the International LBD Genomics Consortium, The American Genome Center, Rosa Rademakers, Clifton L. Dalgard, Bradley F. Boeve, Thomas G. Beach, Dennis W. Dickson, Owen A. Ross, Sonja W. Scholz*.
Our partnership in the AMP® PD effort is a meaningful example of the importance of collaboration, resource generation, and data sharing to accelerate discovery for PD research. Within AMP® PD, researchers can access eight different cohorts with common clinical and genomic data along with GP2 datasets. With each GP2 data release, registration increased on average by 27%.

By supporting open science, we facilitate the rapid and free exchange of scientific ideas, ensuring that the research we fund can be leveraged for future discoveries. Open science is at the heart of ASAP’s mission and work.
From the start, ASAP established progressive open science policies to enable science to go further, faster, and at a greater scale.
SUPPORT COLLABORATION/GENERATE RESOURCES
Actively Engaging with the Broader Open Science Community
- Managing Director Ekemini Riley and Scientific Director Randy Schekman are co-chairing the funders group of the National Academies in Science Engineering and Medicine Funders Roundtable on Aligning Incentives for Open Science.
- Deputy Director Sonya Dumanis was a judge for the 2022 Neuro – Irv and Helga Cooper Foundation Open Science Prizes Open Science Prize.
- ASAP continues to be an active member of both CoalitionS and ORFG subcommittees.
- Resources developed by ASAP are incorporated into the ORFG Open Scholarship Policy Clause Bank and the Open & Equitable Model Funding Program.
GENERATE RESOURCES
Supporting Open-Science Implementation Frameworks
- Our efforts are accelerating the development of an emerging open science tool chain for discovery and impact assessment.
- Our compliance tool, co-developed with DataSeer.Ai, is now being utilized by the Children’s Tumor Foundation as an educational tool for grantees to learn about data sharing as they prepare manuscripts.
- We are piloting a new tool from OA.Works for the discovery of manuscripts authored by ASAP grantees that feed into our DataSeer.Ai system.
- We released the ASAP Blueprint for Collaborative Open Science which provides a roadmap for facilitating open science.

ACCELERATE DISCOVERIES
Pioneering a Path for Open Science
Our efforts to create an open science environment have generated the following outputs:
- Incentivizing Collaborative and Open Research (ICOR) is developing an initiative for a research output management system (ROMS) wiki inspired by our data schema model.
- We are advancing collaborative and transparent research processes and environments to deliver faster and better outcomes to accelerate discovery and inform the path to a cure for PD. The application of this approach—as outlined in the ASAP Blueprint for Collaborative Open Science—extends beyond PD to offer a roadmap for like-minded researchers and funders in support of open science and structuring teams that support collaboration, foster creativity, and improve the efficiency of solution creation. The BD2: Breakthrough Discoveries for thriving with Bipolar Disorder Discovery Grant program adopted a similar model and is facilitating the rapid and free exchange of scientific ideas to reach their goals.
- ORFG is currently working with 11 funders who are implementing variations of ASAP open policy and/or utilizing our open science primers in their own open science initiatives (for example, see Templeton World Charity Foundation).
- ASAP commissioned a study around ORCID utilization within our network from Laura Paglione, who presented in an HRA Webinar, “From Policy to Practice”.
We are excited to be pioneering a path of open science for the ASAP community and the research community at large. Please join our listserv if you want to receive updates on any of our ASAP activities throughout the year.
Thank You to Our Partners
We wish to extend our gratitude to our implementation partner The Michael J. Fox Foundation, and our collaborators Stratos, DataSeer, Parkinson’s Foundation, Parkinson’s UK, and AMP® PD for working with us to implement our vision. Our collective commitment to better understand the underlying causes of PD has allowed us to achieve the above outputs in 2022. We look forward to continuing our efforts in the new year.






A Message From Executive Leadership
We are proud of the progress made in 2022 toward our vision of advancing collaborative, transparent research processes and environments that deliver faster and better outcomes in Parkinson’s disease research. Aligning Science Across Parkinson’s (ASAP) thanks the Sergey Brin Family Foundation for their generous support in funding this initiative, our partners, the research community, and all our stakeholders for the contributions they have made to our research network and the entire neuroscience community. We look forward to our continued partnership and research community efforts, propelled by the success of 2022 and the lessons we learned along the way. Onward to 2023!

