PD Functional Genomics | 2020
Impaired Integration of Organelle Function in Parkinson’s Disease
Study Rationale: Geneticists have made great progress in identifying gene mutations that either cause Parkinson’s or increase disease risk. The critical next step is to determine how some of these mutations perturb the function of brain cells involved in Parkinson’s disease. By advancing knowledge of these processes, Team De Camilli’s research will help to identify new opportunities for reversing the vulnerabilities that cause the disease.
Hypothesis: Team De Camilli hypothesizes that the functions of multiple Parkinson’s disease genes converge on common biochemical pathways involving endocytic organelles and/or mitochondria within vulnerable cell types.
Study Design: Team De Camilli will use a comprehensive cell biology tool kit including cutting-edge biochemistry, structural biology, microscopy at different scales, and genome editing tools to elucidate the function of selected Parkinson’s disease genes and the effects produced by their dysfunction both in cellular models in vitro and in mouse and rat models. By defining the molecular and cellular networks in which the products of these genes operate, Team De Camilli hopes to identify strategies for reversing the cellular vulnerabilities that cause Parkinson’s disease or increase disease risk.
Impact on Diagnosis/Treatment of Parkinson’s Disease: Similar to assembling the pieces of a puzzle, the project has the potential to reveal interconnections between the functions of distinct Parkinson’s disease genes, thus helping to build an understanding of Parkinson’s disease cell biology. This is a critical step towards the development of therapeutic strategies to make neurons resistant to the dysfunctions that cause Parkinson’s disease.
Leadership
Project Outcomes
The project hopes to reveal interconnections between the functions of distinct Parkinson’s disease genes, thus helping to identify cellular processes whose dysfunction confers Parkinson’s disease vulnerability and which may be targeted for therapeutic intervention. View Team Outcomes.
Team Outputs
Click the following icons to learn more about the team’s outputs:
Overall Contributions
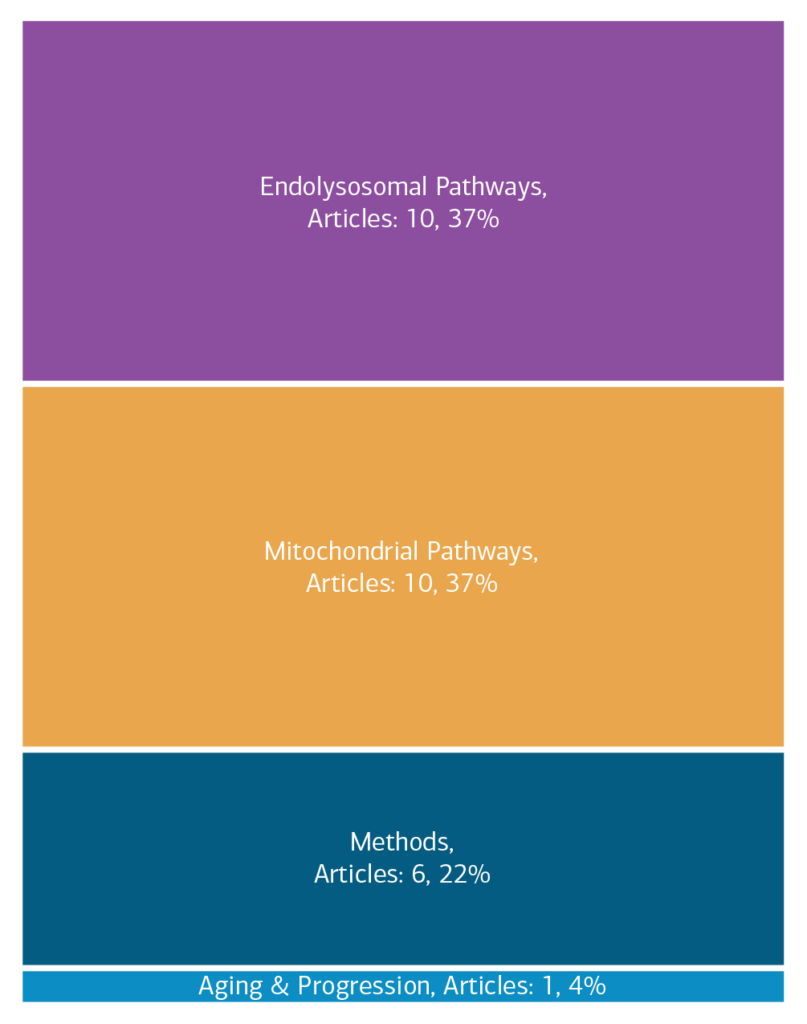
Here is an overview of how this team’s article findings have contributed to the PD field as of November 2023. There are two different categorizations of these contributions – one by impact to the PD community and a second by scientific theme.
Impact

Theme
Featured Output
Below is an example of a research output from the team that contributes to the ASAP mission of accelerating discoveries for PD.
Structural and biochemical insights into lipid transport by VPS13 proteins
Team De Camilli has provided direct evidence for VPS13C serving as a bridge between ER and lysosomes. This discovery paves the way to better understand the role of VPS13C in normal function and the impact of mutations in VP13C in PD dysfunction.
Team Accolades
Members of the team have been recognized for their contributions.
- Open Science Champions: De Camilli Lab
- Network Spotlights: William Hancock-Cerutti
- Awards
- 2023 Collaborative Meeting: Amanda Bentley de Souza, First Place Winner in PD Functional Genomics; London

Other Team Activities
-
Working Groups: Single Cell Multi(Omics) – Le Zhang (Subgroup Lead)
- Interest Groups: Endolysosomal Pathways – Shawn Ferguson (Chair)
In the News
- Yale teams get multi-million-dollar awards to study biology of Parkinson’s (Yale News, press release, September 16, 2020)












